Significancy
Microbial rhodopsins, particularly the channelrhodopsin family of green algae, have come into the focus of neurobiological research as very useful tools to study connections and the functioning of neuronal networks (optogenetics). The light-activated ion channel channelrhodopsin-2 (ChR2) has become the cornerstone of this emerging field. Like all other known microbial rhodopsins, ChR2 spans the membrane in seven transmembrane helices (7-TM), and the chromophore retinal is covalently attached to a lysine via a protonated Schiff base. ChR2 is a powerful tool to study cellular processes with high spatiotemporal resolution in the emerging field of optogenetics. To customize the channel properties for optogenetic experiments, a detailed understanding of its molecular reaction mechanism is essential.
Main Goals
A major challenge for current biophysical research is the understanding of both, detailed functional mechanisms and interactions of membrane proteins, especially of heptahelical membrane-proteins. We focus on microbial rhodopsins, specifically bacteriorhodopsin (bR), Channelrhodopsin 2 (ChR2), and G-protein coupled receptors (GPCRs. Our major interest lies in the interaction between proteins and small molecules, such as retinal and water molecules. Our research group has especially contributed to the understanding of the role of water molecules in protein structure stabilization and protein-internal proton storage and transduction. Our major focus for these studies has been the prototypical protein proton pump and microbial rhodopsin bR. In addition to bacteriorhodopsin the light-gated ion-channel ChR2 that is highly important for neurobiology, has become the major focus of our research. The so-called “optogenetics” have become a promising, fast evolving branch. Optogenetic applications are advantageous, because they are less invasive than other methods and provide a high spatial and temporal resolution in neurologic stimulation. This revolutionary method is based on the usage of genetically targetable photosensitive proteins in order to control the activity of excitable cells by light. Within this field, ChR2, a microbial rhodopsin from Chlamydomonas rheinhardtii, has become a major tool. ChRs in general are members of a specific subclass of sensory rhodopsins that mediate phototaxis and photophobic behavior in flagellate green algea. Despite the variety of applications in neurobiology, little is known about the molecular mechanisms of ion pore formation. In this study, we aim to close this gap by incorporating information we get from biophysical methods such as electrophysiological measurements. time-resolved FTIR-, Raman- and UV/Vis spectroscopy into our Molecular Dynamics simulations.
Protonation changes and pore formation in channelrhodopsins
In our previous publications on ChR2 (see Eisenhauer et al, JBC 2012) we showed that the deprotonation of the glutamic residue E90 causes water influx into the conductive pore. Significant improvements in our FTIR-spectroscopic setup allowed us to monitor the time course of the E90 protonation state with high temporal resolution. We observed an ultrafast deprotonation of E90 no later than 2 µs after start of the reaction. This indicates a direct coupling of E90 to chromophore isomerization. Using an improved ChR2 WT homology model based on the crystal structure of a ChR C1C2 chimera (PDB-ID: 3UG9; Kato et al., Nature 2012) we examined the effect of the isomerization on the hydrogen bonding network involving E90 and N258. In the ground state all-trans retinal configuration E90 on Helix2 is connected to N258 on Helix7 via two hydrogen bonds. To model the all-trans 13-cis isomerization of the retinal we rotated around the C13=C14 double-bond in 20° steps (for details see Kuhne et al. Angew. Chemie Int. Ed. 2015).
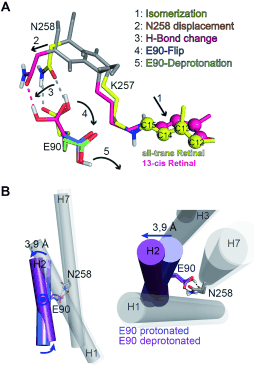
Figure 1:
All-trans to 13-cis isomerization and Helix2 movement in ChR2 based on MD simulations. (A) The side chains are colored and the protein backbone is shown in gray. The all-trans and the fully rotated 13-cis retinal configurations are shown in yellow and magenta. RSBH+ isomerization (1)
is followed by K257 and N258 displacement (2), loss of one hydrogen bond (3), E90 outward flip (4), and E90 deprotonation (5). (B) Movement of Helix2 as a consequence of the events shown in (A). Upon the deprotonation and outward flip of E90, Helix2 is tilted outward by 3.9 Å on the
intracellular side of the protein. E90 within Helix2 works as a hinge for this movement.
We observed an isomerization-induced displacement of N258 which leads to a disruption of the hydrogen bonds to E90. After that E90 flips downward and is able to deprotonate (see Figure 8A). This mechanism explains the ultrafast deprotonation observed in our FTIR experiments. The MD-Simulations further show that after E90 deprotonation an outward tilt of Helix2 occurs with E90 working as a hinge for this movement (see Figure 8B). The observed helix movement is in agreement with ESR-measurements (Krause et al. FEBS Letters 2013). The tilt is caused by water influx into the protein that weakens the hydrogen bonds of the inner gate. Thereby a continuous aqueous pore through the protein, with E90 at the narrowest point, is formed (see Figure 9). The water influx has later been independently confirmed by FTIR measurements of other groups (Lórenz-Fonfría et al. PNAS 2015). This process reflects a pre-formation of the conductive pore that we termed the E90-Helix2-tilt (E-H-T) model (Kuhne et al. Angew. Chemie Int. Ed. 2015).
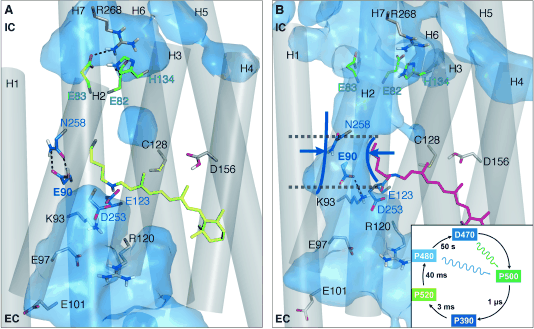
Figure 2:
ChR2 in (A) the closed and (B) preopen state. The isomerization-induced deprotonation of E90 leads to a preopening of the pore. In the closed state the hydrogen bonds between E90 and N258 (central gate) as well as E83 and R268 (inner gate) connect Helix2 with Helix7.
Both connections prevent water influx into the intracellular vestibule. In the preopened state these connections are lost because of the flip of E90 from the inward (IC) to the outward (EC) orientation and water influx into the protein.
Current projects: pore-opening mechanism of Channelrhodopsin 2
Following our earlier research on ChR2, we are currently investigating additional isomerization events which we found in experiments. Our recent Raman and FTIR-spectroscopic measurements show that an early splitting of the photocycle involving an all-trans, C=N-anti 13-cis, C=N-anti and an all-trans, C=N-anti 13-cis, C=N-syn occurs. In future we plan to incorporate these findings into our MD-simulations and analyze the response of the protein.
Medium and Long Term Research Goals
Press Releases
Publications
- Die frühe Entstehung der ionenleitenden Pore in Channelrhodopsin-2 Kuhne, J., Eisenhauer, K., Ritter, E., Hegemann, P., Gerwert, K., Bartl, F.
- Early Formation of the Ion-Conducting Pore in Channelrhodopsin-2 Kuhne, J., Eisenhauer, K., Ritter, E., Hegemann, P., Gerwert, K., Bartl, F.
- Infrared spectral marker bands characterizing a transient water wire inside a hydrophobic membrane protein Wolf, S., Freier, E., Cui, Q., Gerwert, K.
- A Delocalized Proton-Binding Site within a membrane Protein Wolf, S., Freier, E., Gerwert, K.
- The role of protein-bound water molecules in microbial rhodopsins Gerwert, K., Freier, E., Wolf, S.
- In channelrhodopsin-2 E90 is crucial for ion selectivity and is deprotonated during the photocycle Eisenhauer K., Kuhne J., Ritter E., Berndt, A., Wolf S., Freier E., Bartl F., Hegemann P., Gerwert K.
- Prediction of a Ligand-Binding Niche within a Human Olfactory Receptor by Combining Site-Directed Mutagenesis with Dynamic Homology Modeling Gelis, L., Wolf, S., Hatt, H., Neuhaus, E.M., Gerwert, K.
- Vorhersage der Ligandenerkennung in einem Geruchsrezeptor durch Kombination von ortsgerichteter Mutagenese und dynamischer Homologie-Modellierung Gelis, L., Wolf, S., Hatt, H., Neuhaus, E.M., Gerwert, K.
- The structure of active opsin as a basis for identification of GPCR agonists by dynamic homology modelling and virtual screening assays Schneider, M., Wolf, S., Schlitter, J., Gerwert, K.
- Structural Basis of Slow Activation Gating in the Cardiac IKs Channel Complex Strutz-Seebohm, N. Pusch, M., Wolf, S., Stoll, R. Tapken, D., Gerwert, K., Attali, B., Seebohm, G.
- Proton transfer via a transient linear water-molecule chain in a membrane protein Freier, E., Wolf, S., Gerwert, K.
- Directional Proton Transfer in Membrane Proteins Achieved through Protonated Protein-Bound Water Molecules: A Proton Diode Wolf, S., Freier, E., Potschies, M., Hofmann, E., Gerwert, K.
- Gerichteter Protonentransfer in Membranproteinen mittels protonierter proteingebundener Wassermoleküle: eine Protonendiode Wolf, S., Freier, E., Potschies, M., Hofmann, E., Gerwert, K.
- How Does a Membrane Protein Achieve a Vectorial Proton Transfer Via Water Molecules? Wolf, S., Freier, E., Gerwert, K.
- Simulations of a G protein-coupled receptor homology model predict dynamic features and a ligand binding site Wolf, S., Böckmann, M., Höweler, U., Schlitter, J., Gerwert, K.
- Water Dynamics Simulation as a Tool for Probing Proton Transfer Pathways in a Heptahelical Membrane Protein Kandt, C., Gerwert, K., Schlitter, J.
- Computational Studies on Early Intermediates of Bacteriorhodopsin Photocycle Engels, M., Gerwert, K., Bashford, D.
Angew. Chem. 2015, 127, 5037-5041
Angew. Chem. Int. Ed. 2015, 54, 4953–4957
J. Chem. Phys. 141, 2014 22D524
Biophysical Journal 107 2014 174–184
Biochim. Biophys. Acta 1837 2014 606–613
Journal of Biological Chemistry, 287 (9), 2012 6904-6911
Angew. Chem., 51, 2012 1274-1278
Angew. Chem., 124, 2012 1300-1304
FEBS Letters 585, 2011 3587-3592
Cell Physiol Biochem 27, 2011 443-452
PNAS 108 (28), 2011 11435-11439
Angew. Chem. Int. Ed. 49, 2010, 6889-6893
Angew. Chem. 122, 2010, 7041-7046
ChemPhysChem 9(18), 2008, 2772-2778
FEBS Lett. 582(23), 2008 3335-3342
PROTEINS: Structure, Function, and Bioinformatics, 58, 2005 528-537
Biophysical Chemistry 56, 1995 95-104
Cover
Investigators
- Prof. Dr. Klaus Gerwert
- PD Dr. Mathias Lübben
- Dr. Till Rudack
- Dro. Jens Kuhne
- Max Dreier
- Stefan Tennigkeit
- Vincent Walter
- Philipp Althoff
- Felix Diehl
- Christian Benedict Smit

